DNase I
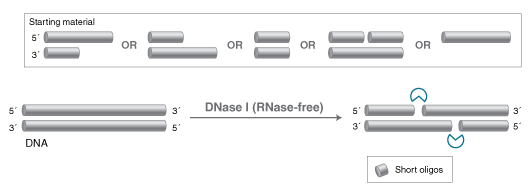
DNase I (deoxyribonuclease I, RNase-free) is a recombinant enzyme that cleaves DNA nonspecifically to release short oligonucleotides with 5' phosphorylated and 3' hydroxylated ends. It is derived from a strain of E. coli that carries the gene for bovine pancreatic DNase I.
DNase I: A Versatile Enzyme with a Wide Range of Applications
DNase I
DNase I is a versatile enzyme that can be used for a variety of molecular biology applications, including:
- DNA digestion: DNase I can be used to digest DNA into smaller fragments for analysis or cloning.
- Removal of RNA contamination: DNase I can be used to remove RNA contamination from DNA samples.
- Nick translation: DNase I can be used to create nicks in DNA strands for labeling or repair reactions.
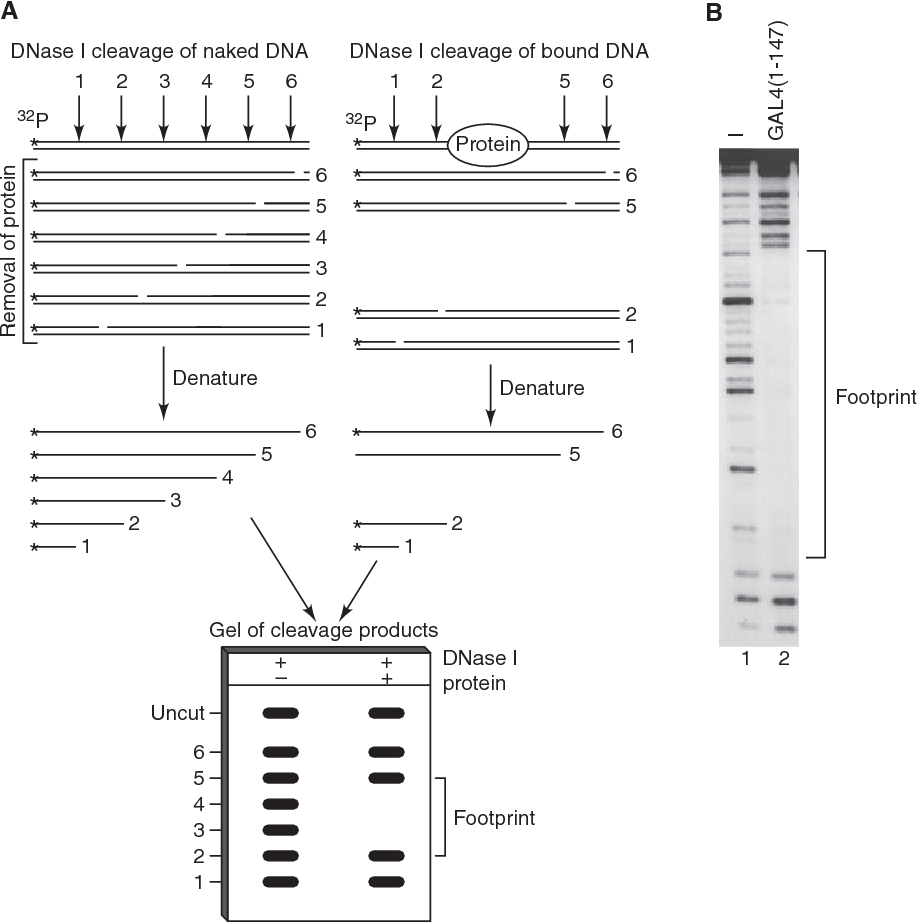
- Footprinting: DNase I can be used to identify protein binding sites in DNA.
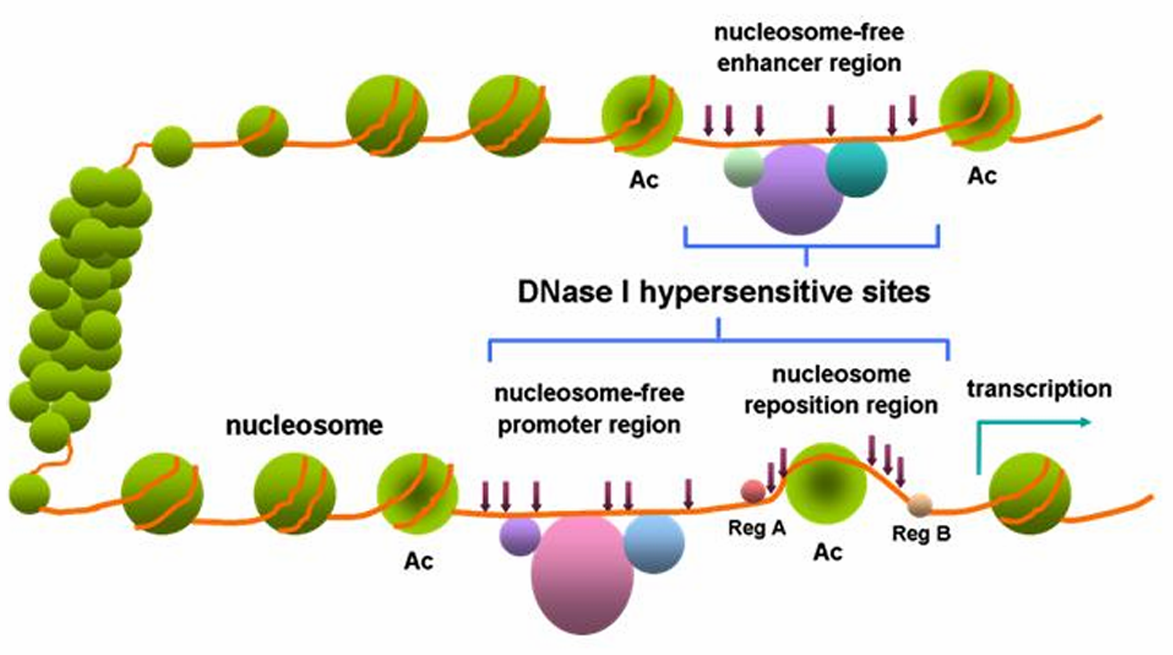
- Chromatin accessibility assays: DNase I can be used to measure the accessibility of chromatin to nucleases, which can be used to study gene expression and regulation.
Features
- DNase I has a relatively high turnover number, meaning that it can cleave a large number of DNA molecules in a short period of time.
- DNase I is relatively stable at high temperatures and pH values, making it a robust enzyme for use in a variety of experimental conditions.
- DNase I is relatively inexpensive and readily available from a variety of commercial sources.
| Feature | Value |
|---|---|
| Type | Endonuclease |
| Activity | Nonspecific |
| Substrate | DNA (single-stranded, double-stranded, and RNA:DNA hybrids) |
| Product | Oligonucleotides with 5' phosphorylated and 3' hydroxylated ends |
| Optimum pH | 7.5-8.0 |
| Optimum temperature | 37°C |
| Requirements | Mg2+ or Ca2+ ions |
| Inhibited by | Copper ions |
| Resistant to | Proteases |
| Purity grades | RNase-free, low-salt |
| Applications | DNA digestion, RNA removal, nick translation, footprinting, chromatin accessibility assays |
Specification
Product component
| Component | Volume (0.1 KU) | Volume (1 KU) | Volume (10 KU) | Volume (100 KU) |
|---|---|---|---|---|
| DNase I (2 U/μL) | 50 μL | 500 μL | 5 mL | 50 mL |
| 10× DNase I Buffer | 100 μL | 1 mL | 10 mL | 100 mL |
Notes:
- The volume of DNase I needed will depend on the desired activity of the enzyme in the reaction. For example, a 10 KU reaction will require 5 mL of DNase I, while a 0.1 KU reaction will only require 50 μL.
- The 10× DNase I Buffer is provided separately from the DNase I enzyme. This allows users to adjust the concentration of the buffer in the reaction as needed.
Example:
To prepare a 10 KU DNase I reaction, you would add the following components to a reaction tube:
- 5 mL DNase I (2 U/μL)
- 10 mL 10× DNase I Buffer
- 100 μL water (or other desired buffer)
The total reaction volume would be 15 mL.
Storage Condition: -20℃ for storage(Avoid repeated freeze-thaw cycles)
Storage Buffer: 2 mM CaCl2,10 mM Tris-HCl (pH 7.6,25℃), 50% glycerol.
Unit Definition: One unit is defined as the amount of enzyme which will completely degrade 1 µg of pBR322 DNA in a total reaction volume of 50 µl in 10 minutes at 37°C. Complete degradation is defined as the reduction of the majority of DNA fragments to tetranucleotides or smaller.
Heat inactivation: 75℃,10 min.
Quality Control:
- RNase: 2 U of DNase I with 1.6 μg MS2 RNA for 16 hours at 37 ℃ yields no degradation as determined by agarose gel electrophoresis.
- E.coli DNA: 2 U of DNase I is screened for the presence of E. coli genomic DNA using TaqMan qPCR with primers specific for the E. coli 16S rRNA locus. The E. coli genomic DNA contamination is ≤1 E. coli genome.
- Bacterial Endotoxin: LAL-test, according to Chinese Pharmacopoeia IV 2020 edition, gel limit test method, general rule (1143). Bacterial endotoxin content should be ≤10 EU/mg.
Notes on use:
- It is recommended to use the enzyme storage buffer for dilution.
- EDTA should be added to a final concentration of 5 mM to protect RNA from being degraded during enzyme inactivation.
- For your safety, please wear lab coat and disposable gloves during operation.
Additional Notes:
- DNase I Sp is a highly active endonuclease that cleaves DNA nonspecifically.
- DNase I Sp is commonly used in molecular biology applications such as DNA digestion, RNA removal, nick translation, footprinting, and chromatin accessibility assays.
- DNase I Sp is a stable enzyme that can be used at a variety of temperatures and pH values.
Application :
DNase I: A Versatile Enzyme for Molecular Biology
DNase I is a powerful enzyme that can be used for a variety of tasks in molecular biology research. It is most commonly used to degrade DNA, but it can also be used to remove contaminating genomic DNA from RNA samples, study protein-DNA interactions, and label DNA.
- Degradation of DNA template in transcription reactions: DNase I can be used to degrade the DNA template after in vitro transcription reactions. This prevents the template from being reused in subsequent reactions, which can improve the purity of the RNA product.
- Removal of contaminating genomic DNA from RNA samples: Genomic DNA can contaminate RNA samples during the isolation process. This can interfere with downstream applications such as cDNA synthesis and RNA sequencing. DNase I can be used to remove contaminating genomic DNA from RNA samples, ensuring that the RNA samples are pure and can be used for accurate and reliable results.
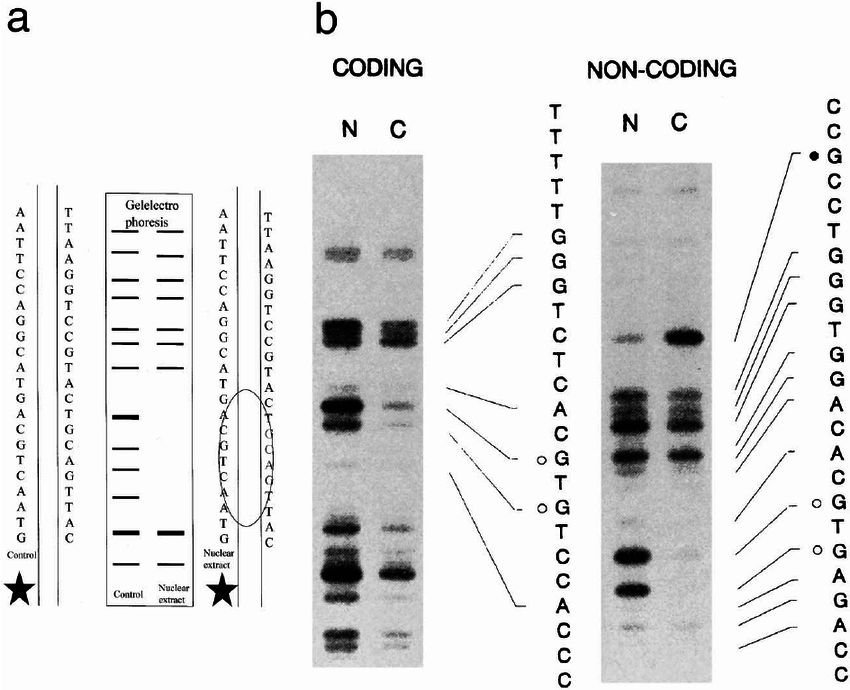
- DNase I foot-printing: DNase I foot-printing is a technique used to study protein-DNA interactions. In this technique, DNase I is used to cleave DNA at sites that are not protected by proteins. The resulting DNA fragments can then be analyzed to identify the regions of DNA that are bound by the protein of interest. DNase I foot-printing is a powerful tool for understanding how proteins interact with DNA, which can lead to new insights into gene regulation and other important biological processes.
- Nick translation: Nick translation is a technique used to label DNA with radioactive or fluorescent nucleotides. This can be useful for a variety of applications, such as hybridization and sequencing. In nick translation, DNase I is used to create nicks in the DNA. DNA polymerase I then extends these nicks using labeled nucleotides. Nick translation can be used to label DNA probes, which can then be used to detect specific sequences of DNA in a sample.
DNase I is a versatile enzyme that is used in a variety of molecular biology research applications. It is a valuable tool for researchers who are studying DNA, RNA, and protein-DNA interactions.
DNase I, a cornerstone enzyme in molecular biology, plays a pivotal role in DNA research by selectively cleaving phosphodiester linkages within the DNA backbone. This endonuclease, derived from bacteria or fungi, is characterized by its ability to hydrolyze both single-stranded and double-stranded DNA, resulting in the generation of DNA fragments with defined sizes. Researchers employ DNase I for a myriad of applications, ranging from DNA footprinting and chromatin analysis to the removal of DNA contaminants in RNA preparations. The enzyme's preference for cleaving DNA at sites with accessible phosphate groups makes it a valuable tool for investigating chromatin structure, protein-DNA interactions, and studying the spatial organization of genes. Its broad versatility, coupled with the precision it offers in DNA digestion, positions DNase I as an indispensable asset in the molecular biologist's toolkit, contributing significantly to our understanding of genomic structure and function.