Library Preparation Kit for Illumina & MGI
RNA library prep kit
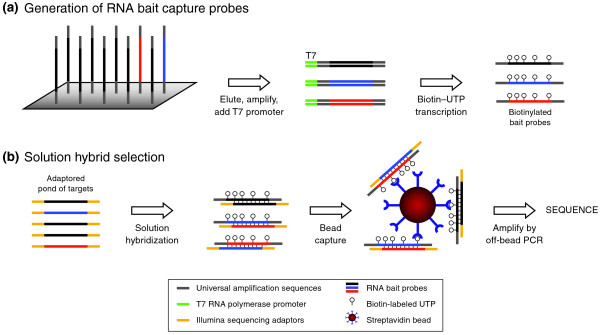
- An RNA library preparation kit enables the construction of high-quality RNA libraries for downstream sequencing analysis. Here is an overview of the process and key steps involved in using an RNA library prep kit:
- RNA Isolation and Quality Assessment:
- Begin with the isolation of high-quality RNA from the biological sample of interest.
- Assess the integrity and purity of the RNA to ensure it is suitable for library preparation.
- RNA Fragmentation:
- Fragment the RNA into smaller pieces using enzymes or other fragmentation methods included in the kit.
- Ensure controlled and uniform fragmentation to facilitate accurate library construction.
- cDNA Synthesis:
- Perform reverse transcription (cDNA synthesis) using the fragmented RNA as a template.
- Incorporate unique barcodes or indexes during cDNA synthesis to enable multiplexing and sample identification.
- End Repair and Adenylation:
- Prepare the cDNA fragments for adapter ligation by conducting end repair and adenylation steps.
- These processes enable the attachment of sequencing adapters to the cDNA fragments.
- Adapter Ligation:
- Ligating sequencing adapters to the cDNA fragments allows for the subsequent amplification and sequencing steps.
- Library Amplification:
- Use PCR or other amplification techniques to enrich the RNA library, making it suitable for sequencing.
- Quality Control:
- Conduct quality control checks to assess the quality and concentration of the RNA library.
- Verify the size distribution of the library fragments to ensure the integrity of the sequencing results.
- Sequencing:
- Follow standard protocols for sequencing on the appropriate platform.
- Analyze the resulting sequence data using relevant bioinformatics tools to extract meaningful insights.
Conventional DNA library prep kit
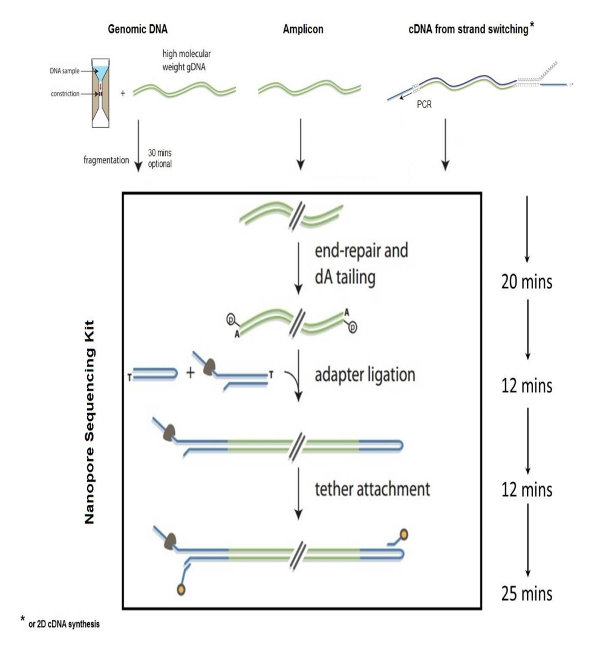
Conventional DNA library prep kits convert fragmented DNA for high-throughput sequencing, supporting diverse genomic studies and offering detailed user manuals for precise library construction.
Enzymatic DNA fragmentation library prep kits
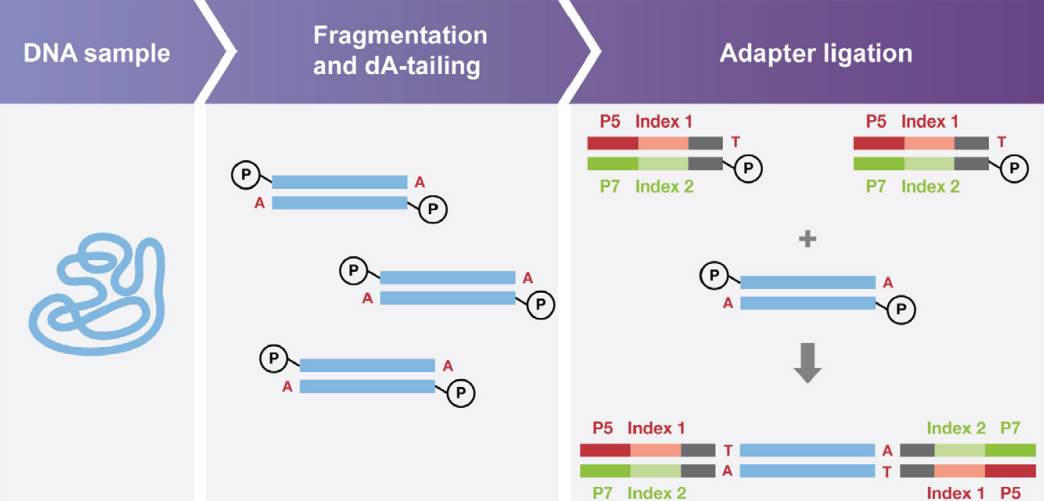
Sample preparation requires high-quality genomic DNA for enzymatic fragmentation, followed by end repair, A-tailing, adapter ligation, library amplification, quality control, and sequencing using an enzymatic DNA fragmentation library prep kit.
Library Preparation Module
Dual barcode cyclization kit
The dual barcode cyclization kit is a specialized tool for facilitating the cyclization of DNA fragments in preparation for targeted sequencing applications, ensuring precise and controlled ligation of sequencing adapters. It offers enhanced accuracy and efficiency in generating high-quality libraries and enables comprehensive sample tracking through the use of dual barcoding systems.
Plant rRNA
removal
Plant rRNA removal refers to the process of selectively depleting ribosomal RNA (rRNA) molecules from plant samples, allowing for enhanced sensitivity and accuracy in downstream RNA sequencing analyses. This technique effectively minimizes the presence of highly abundant rRNA, enabling researchers to focus on rare or low-abundance transcripts of interest and obtain a more comprehensive view of the plant transcriptome.
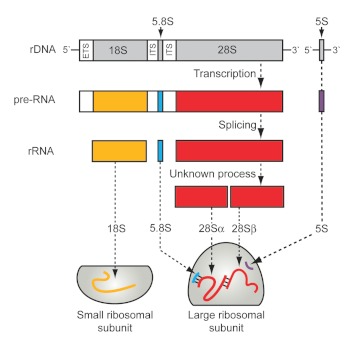
Human rRNA&ITS/ETS removal
Human rRNA & ITS/ETS removal involves the selective elimination of ribosomal RNA and internal transcribed spacer (ITS) or external transcribed spacer (ETS) regions from human RNA samples. This process is essential for enhancing the sensitivity and accuracy of downstream RNA sequencing, allowing for improved detection of low-abundance and biologically significant transcripts, thus providing a more comprehensive understanding of the human transcriptome.
Magnetics Beads

DNA cleanup & size selection
DNA cleanup and size selection are critical steps in the genomic library preparation process that involve the removal of unwanted contaminants and the isolation of specific DNA fragment sizes. These procedures contribute to the generation of high-quality DNA libraries suitable for downstream sequencing applications, ensuring accurate and reliable data analysis and interpretation.

RNA cleanup
RNA cleanup is a crucial process that involves the removal of impurities and contaminants from RNA samples, ensuring the purification and concentration of high-quality RNA for subsequent downstream applications such as reverse transcription, RNA sequencing, and other molecular biology techniques.

mRNA isolation
mRNA isolation is a specialized procedure that involves the extraction and purification of messenger RNA (mRNA) molecules from a biological sample, often using techniques such as oligo-dT affinity purification or poly-A tail enrichment. This process enables researchers to focus specifically on the transcriptome and study gene expression levels in various biological systems.
Adapters & Primers for Illumina
To effectively utilize the Unique Dual Index Primer (Plate), it is crucial to meticulously follow the prescribed procedures. Begin by ensuring a smooth primer plate setup, guaranteeing the absence of any contaminants and appropriate resuspension if necessary. Prepare the samples for sequencing with the required concentration and purity levels. Next, add the designated primer from the plate to each sample, ensuring accurate indexing and thorough mixing. Conduct PCR amplification adhering to the recommended cycling conditions for optimal results. Subsequently, evaluate the quality and quantity of the amplified products through a suitable analytical method, verifying the success of the indexing and amplification steps. Finally, initiate the sequencing process for the prepared samples on the designated platform, followed by a comprehensive analysis of the resulting data using suitable bioinformatics tools to extract valuable insights and interpretations.
Combinatorial Dual Index Primer
The Combinatorial Dual Index Primer is a critical component used in DNA sequencing for the labeling and identification of different samples. It is essential for maintaining the integrity and identity of each sample during the sequencing process. This primer is designed with specific sequences that enable the accurate and efficient indexing of multiple samples simultaneously, allowing for high-throughput sequencing and streamlined data analysis. Its robust design and compatibility with various sequencing platforms make it an essential tool for researchers and scientists in diverse genomic studies.

Adapters & Primers for MGI
Adapters & Primers for MGI offer comprehensive support for targeted sequencing applications, allowing for efficient sample preparation and streamlined sequencing processes. With high-quality components and optimized designs, they ensure accurate and reliable data generation for a wide range of genomic studies.
Enhanced Sample ID
The Unique Dual Barcode Primer (Plate) ensures accurate data retrieval in high-throughput sequencing with distinct barcode sequences for each sample.
Streamlined Multiplexing
Designed for efficient sample processing, it enables simultaneous handling of multiple samples, saving time and maintaining sequencing precision.
Versatile Compatibility
Compatible with diverse sequencing platforms, it offers adaptability to various genomic studies, allowing researchers to explore a wide range of genetic information.
Reliable Data Integrity
With robust barcode sequences, it guarantees consistent and dependable results, ensuring data integrity throughout the sequencing process.