Exploring the Frontiers of Molecular Biology
Molecular Biology
PCR
Polymerase Chain Reaction (PCR) stands as a revolutionary technique in molecular biology, providing researchers with a powerful tool for amplifying specific DNA sequences with unparalleled precision and efficiency. This method, conceptualized by Kary B. Mullis in 1983, has since become a cornerstone in various scientific disciplines, enabling the exponential replication of targeted DNA regions.
At its core, PCR involves a series of temperature-mediated cycles, each comprising denaturation, annealing, and extension steps. Denaturation separates the DNA strands, annealing allows primers to bind to the target sequence, and extension involves the synthesis of new DNA strands by a heat-stable DNA polymerase, often Taq polymerase. The result is an exponential increase in the quantity of the target DNA, making it amenable to subsequent analyses.
PCR's versatility extends across applications such as DNA cloning, genetic sequencing, and diagnostics. Its impact on fields like forensics, medicine, and biotechnology is profound, enabling the study of genetic variations, identification of pathogens, and production of ample DNA for analysis.
Moreover, advancements in PCR technology, including quantitative PCR (qPCR) and reverse transcription PCR (RT-PCR), have expanded its capabilities. QPCR allows precise quantification of DNA, while RT-PCR facilitates the study of gene expression by amplifying complementary DNA (cDNA) from RNA templates.
In essence, PCR has transformed the landscape of molecular biology, facilitating groundbreaking research and applications that were once unimaginable. Its adaptability, efficiency, and reproducibility continue to drive scientific discovery, making PCR an indispensable tool for researchers unraveling the intricacies of the genetic code.
| Classification | Product Name | Cat.No | Size | List Price/$ |
|---|---|---|---|---|
| High Fidelity PCR | 2×Hieff CanaceTM Plus PCR Master Mix (With Dye) | 10154ES03 10154ES08 | 1 mL 5×1mL | 120 336 |
| Conventional PCR | 2×Hieff™ PCR Master Mix (With Dye) | 10102ES03 10102ES08 | 1 mL 5×1 mL | 18 78 |
| 2×Hieff™ PCR Master Mix (No Dye) | 10103ES03 10103ES08 | 1 mL 5×1 mL | 18 78 | |
| Fast PCR | 2×Hieff™ Ultra-Rapid HotStart PCR Master Mix (with Dye) | 10157ES03 | 1 mL | 48 |
| Direct PCR | Mouse Tissue Direct PCR Kit (With Dye) | 10185ES50 10185ES70 | 50 T 200 T | 78 282 |
| dNTP | dNTP Set Solution (dATP, dCTP, dTTP, dGTP, 100 mM each) | 10122ES74 | 4×400 μL | 114 |
| dNTP dNTP Mix (25 mM each) | 10125ES80 10125ES86 | 1 mL 25 mL | 594 14850 | |
| dUTP Solution (100 mM) | 10128ES74 10128ES80 | 400 μL 1 mL | 60 144 |
Cloning
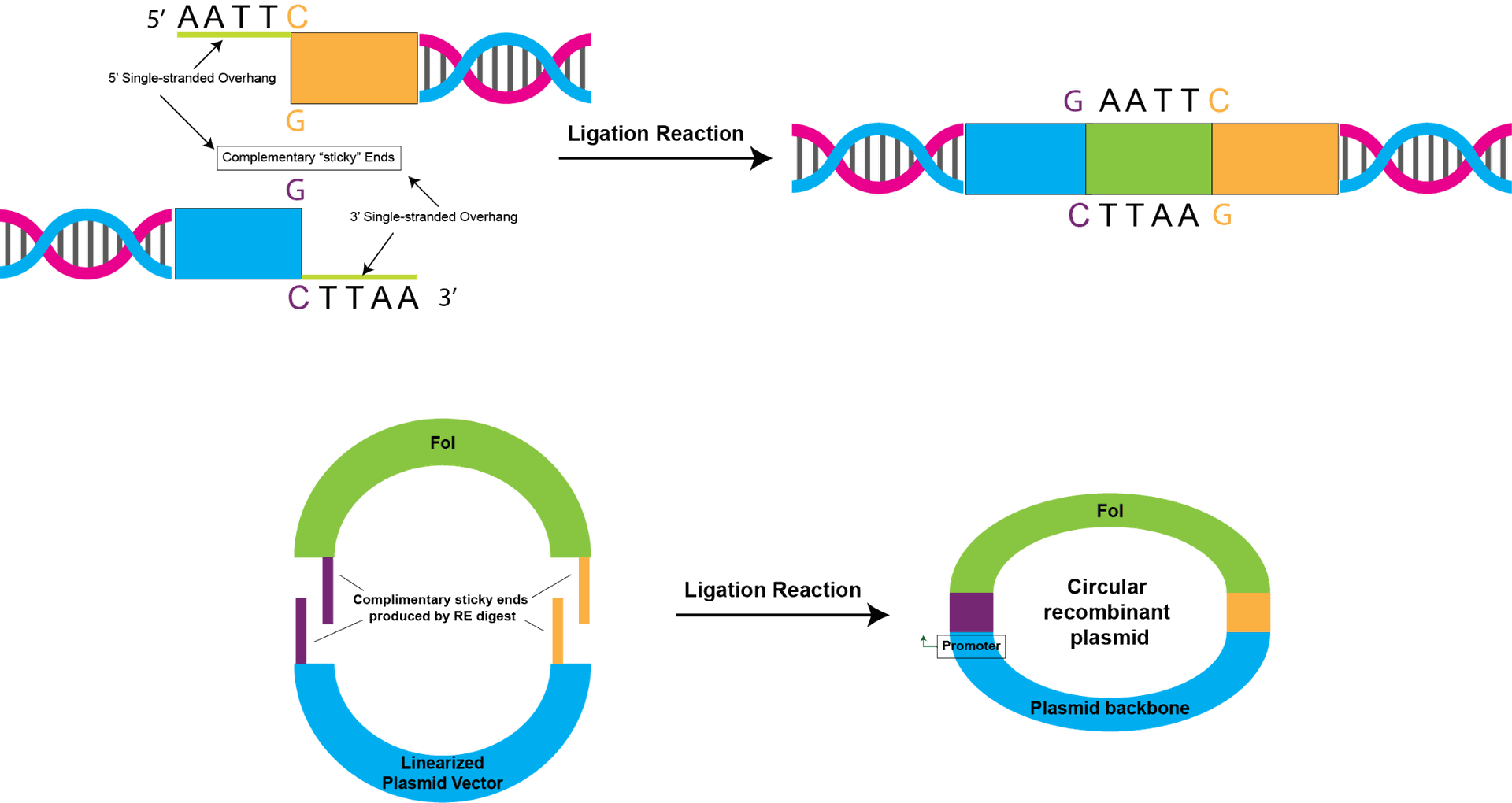
Cloning in molecular biology represents a fundamental technique with profound implications for understanding genetic processes and advancing various scientific endeavors. The term "cloning" refers to the replication of a specific DNA sequence or an entire organism, and in the molecular biology context, it primarily involves the generation of identical copies of a DNA fragment.
In molecular cloning, a target DNA fragment is inserted into a vector, often a plasmid or a viral genome, using specialized enzymes like restriction endonucleases and DNA ligase. The resulting recombinant DNA is then introduced into a host organism, such as bacteria or yeast, where it can be replicated and propagated. This process allows researchers to amplify and study specific genes or DNA sequences of interest.
The applications of molecular cloning are diverse, ranging from gene manipulation and expression to the creation of genetically modified organisms (GMOs) with desired traits. Recombinant DNA technology, an integral part of molecular cloning, has played a pivotal role in producing therapeutic proteins, developing vaccines, and advancing our understanding of gene function.
Furthermore, molecular cloning serves as the foundation for constructing genomic libraries, which are comprehensive collections of cloned DNA fragments representing an organism's entire genome. These libraries provide researchers with a valuable resource for studying genes, their regulatory elements, and their functions.
In summary, cloning in molecular biology is a versatile and indispensable tool that empowers researchers to explore the intricacies of genetics, contributing significantly to scientific progress and applications across diverse fields, from medicine to agriculture.
| Classification | Product Name | Cat.No | Size | List Price/$ |
|---|---|---|---|---|
| Seamless Cloning | Hieff Clone™ Universal One Step Cloning Kit | 10922ES20 10922ES50 | 20 T 50 T | 246 414 |
| TOPO Cloning | Hieff Clone™ Zero TOPO-TA Cloning Kit | 10907ES20 | 20 T | 282 |
| Ligase | 10300ES80 | 1000 U | 66 | |
| Competent Cell | TOP10 Chemically Competent Cell Top10 | 11801ES80 11801ES92 | 10×100 μL 100×100 μL | 408 3060 |
| BL21 (DE3) Chemically Competent Cell BL21 | 11804ES80 | 10×100 μL | 402 | |
| DH5α Fast Chemically Competent Cell F | 11803ES80 | 10×100 μL | 156 | |
| DH5α Chemically Competent Cell DH5α | 11802ES80 11802ES92 | 10×100 μL 100×100 μL | 138 1032 |

https://www.yeabio.com/pages/distributors
Nucleic Acid Electrophoresis
Nucleic Acid Electrophoresis is a crucial analytical technique in molecular biology that enables the separation and visualization of nucleic acids, such as DNA and RNA, based on their size and charge. This method takes advantage of the fact that negatively charged nucleic acids migrate through a gel matrix when an electric field is applied.
The process typically involves loading a sample into wells within an agarose or polyacrylamide gel and applying an electric current. The nucleic acids move through the gel at rates determined by their size, with smaller fragments traveling faster than larger ones. Post-electrophoresis, the gel is often stained with a dye that binds specifically to nucleic acids, allowing for their visualization under ultraviolet light.
Nucleic Acid Electrophoresis serves various purposes in molecular biology. It is a staple technique for verifying the success of DNA extraction, assessing the purity and integrity of DNA or RNA samples, and determining the size of DNA fragments generated during processes like PCR or DNA digestion. Additionally, this method is integral to constructing and analyzing DNA or RNA molecular weight markers.
The versatility of Nucleic Acid Electrophoresis extends to different variants, including agarose gel electrophoresis for separating larger DNA fragments and polyacrylamide gel electrophoresis for resolving smaller fragments with higher resolution. Moreover, capillary electrophoresis has emerged as a more advanced technique, allowing for rapid and automated separation of nucleic acids.
In essence, Nucleic Acid Electrophoresis remains a cornerstone technique in molecular biology laboratories, providing researchers with a robust means of characterizing nucleic acids and contributing to advancements in genetic research, diagnostics, and biotechnology.
| Classification | Product Name | Cat.No | Size | List Price/$ |
|---|---|---|---|---|
| Stain | YeaRed Nucleic Acid Gel Stain (10,000× in Water) | 10202ES30 10202ES76 | 30 μL 500 μL | 18 156 |
| Agarose | Agarose | 10208ES60 10208ES76 | 100 g 500 g | 264 1188 |
| Marker | GoldBand DL2,000 DNA Marker | 10501ES60 10501ES80 | 100 T 10×100 T | 48 336 |
| Marker | GoldBand 5000 DNA Marker | 10504ES60 10504ES80 | 100 T 10×100 T | 48 336 |
| Marker | GoldBand 100bp DNA ladder | 10507ES60 10507ES80 | 100 T 10×100 T | 60 420 |
| Marker | GoldBand 1kb DNA ladder | 10510ES60 10510ES80 | 100T 10×100T | 54 378 |
Reverse Transcription
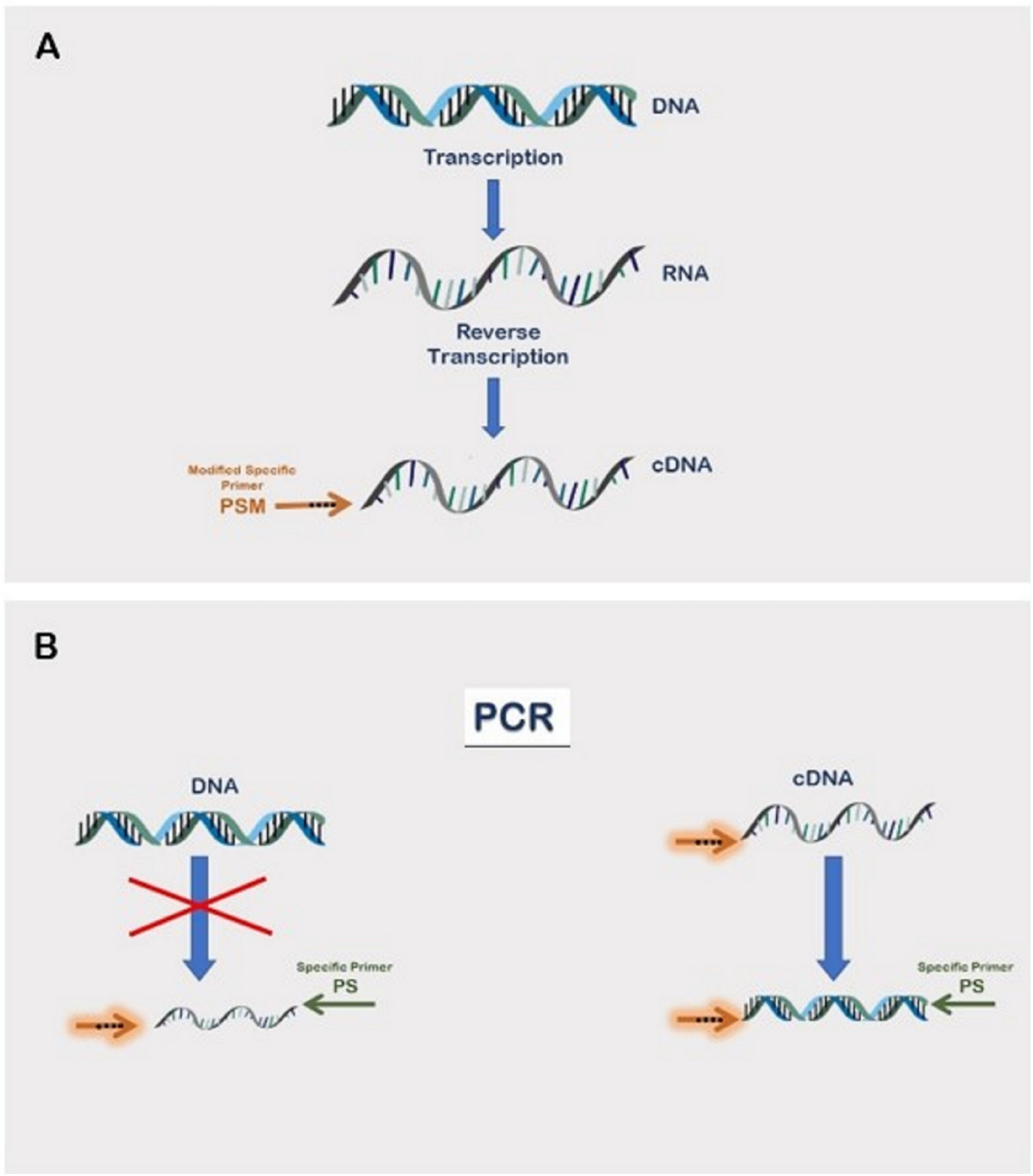
Reverse Transcription is a pivotal molecular biology technique that plays a central role in converting RNA into complementary DNA (cDNA). This process is crucial for various applications, particularly in the study of gene expression and the generation of cDNA libraries.
The process involves the enzyme reverse transcriptase, which synthesizes a complementary DNA strand from an RNA template. This enzymatic activity allows researchers to obtain a DNA copy of specific RNA molecules, providing a means to analyze gene expression levels and study RNA sequences. Reverse transcription is a key component in techniques such as reverse transcription polymerase chain reaction (RT-PCR), quantitative reverse transcription PCR (qRT-PCR), and cDNA library construction.
In RT-PCR, the cDNA generated by reverse transcription serves as a template for subsequent PCR amplification. This enables the quantification of RNA targets, allowing researchers to analyze gene expression levels under different conditions. qRT-PCR further refines this process by providing quantitative data on the initial amount of RNA, offering insights into gene expression dynamics.
Reverse Transcription also plays a critical role in the construction of cDNA libraries, where the cDNA copies of RNA molecules are inserted into plasmids or vectors. These libraries serve as valuable resources for studying gene expression patterns, identifying novel genes, and understanding the complexity of eukaryotic genomes.
The versatility of reverse transcription extends to its applications in virology, where it is essential for replicating the genetic material of RNA viruses, which use RNA as their genomic material. Overall, reverse transcription stands as an indispensable tool in molecular biology, facilitating research across diverse fields and contributing significantly to our understanding of gene expression and RNA biology.
| Classification | Product Name | Cat.No | Size | List Price/$ |
|---|---|---|---|---|
| Individual Random Primers/ Oligo (dT) 18 Primer | Hifair™ II 1st Strand cDNA Synthesis Kit (gDNA digester plus) | 11121ES60 | 100 T | 396 |
| Random Primers/Oligo (dT)18 Primer in the Mix | Hifair™ III 1st Strand cDNA Synthesis SuperMix for qPCR (gDNA digester plus) | 11141ES10 11141ES60 | 10 T 100 T | 30 276 |
| For miRNA | Hifair™ miRNA 1st Strand cDNA Synthesis Kit (poly A) | 11148ES10 11148ES50 | 10 T 50 T | 66 282 |
| Reverse Transcriptase | Hifair™ V Reverse Transcriptase | 11300ES92 11300ES93 | 10000 U 5x10000 U | 456 1836 |
Isothermal Amplification
Isothermal Amplification is a transformative molecular biology technique that allows the rapid and efficient amplification of nucleic acid sequences under constant temperature conditions, eliminating the need for the thermal cycling used in traditional PCR. This method addresses the limitations of conventional PCR and has significant implications for various applications, including diagnostics, point-of-care testing, and field-based analyses.
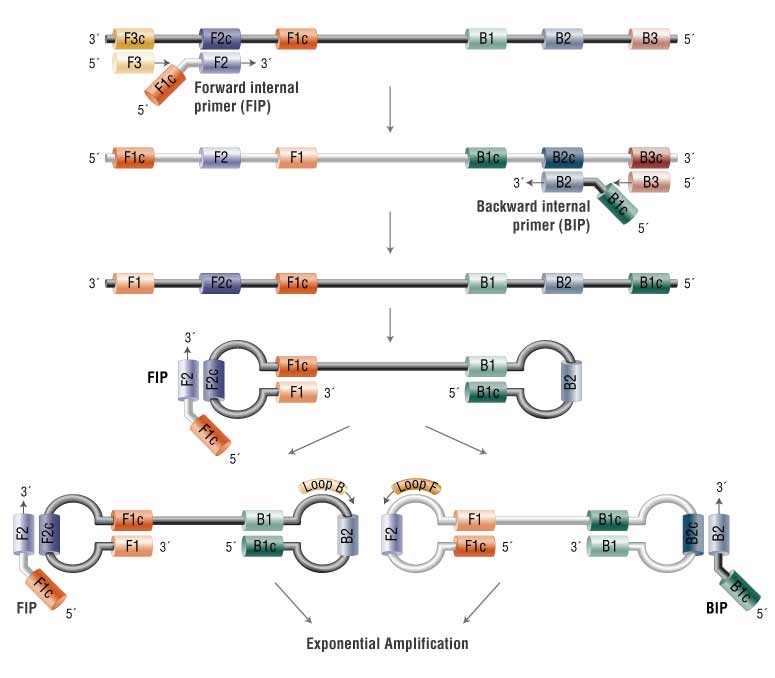
One of the prominent isothermal amplification techniques is Loop-Mediated Isothermal Amplification (LAMP), characterized by its simplicity, speed, and high specificity. LAMP utilizes a set of primers that recognize multiple target sequences, along with a DNA polymerase with strand displacement activity. The amplification reaction occurs at a single, constant temperature, usually between 60 and 65 degrees Celsius.
The LAMP reaction generates a unique stem-loop DNA structure, leading to a distinctive amplification pattern that can be visually inspected. This feature makes LAMP particularly well-suited for applications in resource-limited settings, as it enables results to be interpreted without the need for sophisticated equipment.
Isothermal amplification techniques, including LAMP, are instrumental in nucleic acid detection, offering advantages such as rapid turnaround times, minimal equipment requirements, and increased tolerance to sample inhibitors. These methods have been pivotal in the development of rapid diagnostic tests for infectious diseases, including viral and bacterial infections.
Moreover, the isothermal amplification approach extends beyond LAMP to include other methods like Recombinase Polymerase Amplification (RPA) and Helicase-Dependent Amplification (HDA), each with its unique advantages and applications.
In essence, Isothermal Amplification represents a paradigm shift in nucleic acid amplification technology, opening up new possibilities for sensitive and rapid molecular diagnostics in diverse settings, from clinical laboratories to field-based testing environments. Its efficiency, simplicity, and versatility position isothermal amplification as a valuable tool for advancing molecular biology applications and addressing the evolving needs of diagnostics and research.
| Classification | Product Name | Cat.No | Size | List Price/$ |
|---|---|---|---|---|
| Dye Assay | RT-LAMP Dye Assay Kit (UDGplus) | 13762ES60 | 100 T | 384 |
| Polymerase | HieffTM Bst Plus DNA Polymerase (40 U/μL) | 14402ES92 14402ES97 | 8000 U 40000 U | 66 282 |
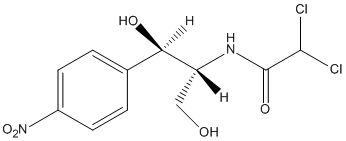
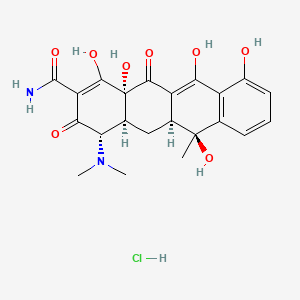
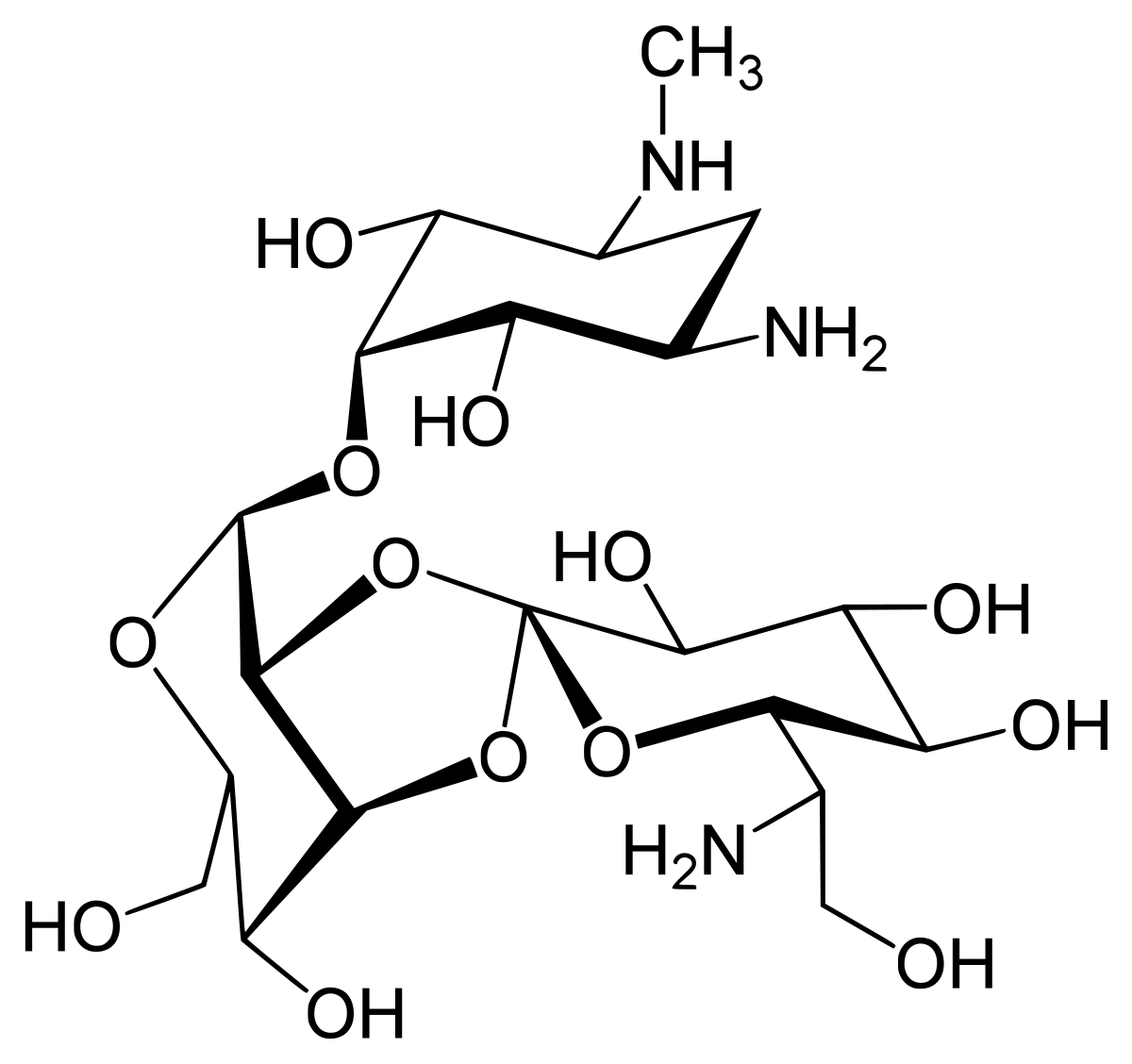
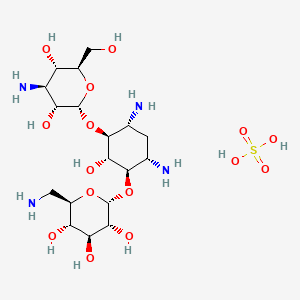
Antibiotics
Ampicillin Sodium Salt
Supplier:Gentaur
Ampicillin Sodium Salt is commonly used in molecular biology and microbiology research as an antibiotic selection agent. To use Ampicillin Sodium Salt, follow these general guidelines:
- Preparation of Ampicillin Solution:
- Prepare a stock solution of Ampicillin Sodium Salt by dissolving the appropriate amount in distilled water or an appropriate buffer.
- Sterilize the solution by passing it through a sterile filter or by autoclaving.
- Store the solution at the recommended temperature (usually -20°C for long-term storage or 4°C for short-term storage).
- Application:
- Ampicillin is often used in bacterial culture media to select for cells containing plasmids with the ampicillin resistance gene.
- Add the appropriate amount of Ampicillin solution to your culture medium (usually in the range of 50-100 μg/mL for E. coli) before pouring plates or inoculating liquid cultures.
- Ensure the agar plates or liquid culture medium are mixed thoroughly to ensure an even distribution of the antibiotic.
- Procedure:
- Prepare your bacterial culture or transform competent cells with your plasmid of interest.
- Plate the transformed cells on solid agar plates containing the appropriate concentration of Ampicillin.
- Incubate the plates at the optimal temperature for your bacterial strain (usually 37°C for E. coli).
- Examine the plates after the appropriate incubation period to determine the growth of colonies, confirming the successful transformation of the plasmid.
G418 Sulfate, commonly known as Geneticin, is an aminoglycoside antibiotic used in genetic selection experiments. When using G418 Sulfate, follow these general instructions and techniques:
- Preparation of G418 Solution:
- Prepare the G418 stock solution by dissolving the appropriate amount of G418 Sulfate in a suitable buffer or growth medium.
- Ensure the solution is thoroughly mixed to achieve a uniform concentration.
- Application:
- G418 Sulfate is commonly used as a selective agent in cell culture experiments to identify and maintain cells that express the neomycin resistance gene (neo) or related genetic markers.
- Add the G418 Sulfate solution to the growth medium to select for cells that contain the gene of interest.
- Techniques and Procedures:
- Determine the optimal concentration of G418 Sulfate required for the selection of cells with the appropriate genetic marker.
- Monitor the cells regularly to assess their response to G418 treatment, focusing on factors such as cell viability, growth rate, and the appearance of resistant colonies.
- Utilize appropriate control cells to accurately gauge the impact of G418 on your experimental system.
Chloramphenicol, USP Grade
Supplier:Gentaur
Cut:100 g
Chloramphenicol, USP Grade, is a broad-spectrum antibiotic commonly used in microbiology and molecular biology research. To use Chloramphenicol, follow these general guidelines:
- Preparation of Chloramphenicol Solution:
- Dissolve the appropriate amount of Chloramphenicol, USP Grade, in a suitable solvent or buffer, typically ethanol or methanol, to create a stock solution.
- Sterilize the solution by filtration using a sterile filter, or by autoclaving if the solvent allows.
- Store the Chloramphenicol solution at the recommended temperature, typically -20°C for long-term storage or 4°C for short-term storage.
- Application:
- Chloramphenicol is commonly used for the selection of bacteria carrying recombinant plasmids that contain the chloramphenicol resistance gene.
- Add the appropriate amount of Chloramphenicol solution to your culture medium before pouring plates or inoculating liquid cultures.
- Ensure the agar plates or liquid culture medium are mixed thoroughly to ensure an even distribution of the antibiotic.
- Procedure:
- Prepare your bacterial culture or transform competent cells with your plasmid of interest.
- Plate the transformed cells on solid agar plates containing the appropriate concentration of Chloramphenicol (usually 10-25 μg/mL).
- Incubate the plates at the appropriate temperature for your bacterial strain (typically 37°C for E. coli).
- Examine the plates after the specified incubation period to assess the growth of colonies, indicating the successful transformation of the plasmid.
Tetracycline hydrochloride (Tetracycline HC)
Tetracycline hydrochloride (Tetracycline HC) is a broad-spectrum antibiotic commonly used in molecular biology and cell culture studies. To use Tetracycline HC effectively, follow these general guidelines and techniques:
- Preparation of Tetracycline Solution:
- Prepare the Tetracycline stock solution by dissolving the appropriate amount of Tetracycline in a suitable solvent or buffer.
- Ensure the solution is thoroughly mixed to achieve a uniform concentration.
- Application:
- Tetracycline is often used to induce or repress gene expression in bacterial cultures or stable cell lines containing specific Tetracycline-responsive elements.
- Add the Tetracycline solution to the culture medium to initiate the desired regulatory effect on gene expression.
- Techniques and Procedures:
- Ensure that you are familiar with the concentration range and exposure time required for your specific application.
- Monitor the response of the cells or organisms to Tetracycline treatment, considering factors such as growth rate, gene expression, and desired experimental outcomes.
- Employ suitable controls to accurately assess the effects of Tetracycline on your experimental system.
Puromycin is an aminonucleoside antibiotic that is commonly used as a selective agent in molecular and cell biology studies. When working with Puromycin (Solution 10 mg/mL), follow these general guidelines:
- Preparation of Puromycin Solution:
- Dilute the Puromycin stock solution to the desired concentration using an appropriate buffer or medium. Typical working concentrations range between 1 to 10 μg/mL for most applications.
- Ensure the solution is thoroughly mixed to achieve a uniform concentration.
- Application:
- Puromycin is used to select for cells that express the antibiotic resistance gene, typically in the context of stable cell lines or during transient transfection experiments.
- Add the appropriate amount of Puromycin solution to the culture medium to allow for the selection and maintenance of cells expressing the resistance gene.
- Procedure:
- Seed your cells of interest in culture plates or flasks according to your experimental requirements.
- Add the Puromycin solution to the cell culture medium to achieve the desired concentration, ensuring that the cells are continuously exposed to the antibiotic.
- Monitor the cells regularly to assess their response to Puromycin, and observe the death or survival of cells lacking the resistance gene.
Hygromycin B
Hygromycin B is an aminoglycoside antibiotic commonly used for the selection of cells carrying the hygromycin resistance gene. When using Hygromycin B, follow these general instructions and techniques:
- Preparation of Hygromycin Solution:
- Prepare the Hygromycin B stock solution by dissolving the appropriate amount of the antibiotic in a suitable buffer or growth medium.
- Ensure the solution is thoroughly mixed to achieve a uniform concentration.
- Application:
- Use Hygromycin B as a selective agent in cell culture experiments to identify and maintain cells containing the hygromycin resistance gene or related markers.
- Add the Hygromycin B solution to the growth medium to select for cells that carry the gene of interest.
- Techniques and Procedures:
- Determine the optimal concentration of Hygromycin B needed for the selection of cells with the appropriate genetic marker.
- Regularly monitor the cells to assess their response to Hygromycin B treatment, focusing on factors such as cell viability, growth rate, and the appearance of resistant colonies.
- Employ suitable control cells to accurately evaluate the impact of Hygromycin B on your experimental system.