Discover the World of Single-Cell Sequencing with Gentaur
what is single cell sequencing ?
Single-cell sequencing is a cutting-edge molecular biology technique that allows researchers to analyze the genetic and genomic information of individual cells within a complex biological sample. Unlike traditional sequencing methods that provide information about the average characteristics of a cell population, single-cell sequencing offers insights into the genetic and functional diversamong individual cells.
This technology has revolutionized our understanding of cellular heterogeneity in various biological contexts, such as cancer research, neuroscience, immunology, and developmental biology. It enables the identification of rare cell types, the study of cell-to-cell variability, and the discovery of novel cell subpopulations. Single-cell sequencing involves the isolation and sequencing of DNA or RNA from individual cells, and it can provide detailed information about gene expression, mutation analysis, epigenetics, and more at a single-cell level. This powerful tool has broad applications in both basic research and clinical studies, offering new perspectives and opportunities for breakthrough discoveries.

"Gentaur Molecular" offers a wide variety of products for single-cell sequencing
1. Single Cell 3' Transcriptome kit
SeekOne DD (Digital Droplet) Single Cell 3' Transcriptome-seq is a high-throughput single-cell RNA-seq library kit achieves single-cell partitioning and labelling by the microfluidic system and barcoded beads. The kit needs to be used with the SeekOneDigital Droplet System (SeekOneDD) to complete the whole process from single-cell transcriptome barcoding to library construction.
SeekOne DD single cell 3' transcriptome kit includes: SeekOne DD Chip S3 (Chip S3 for short), Gasket, Carrier Oil, SeekOne DD 3' Barcoded Beads (Barcoded Beads for short), amplification reagents, library construction reagents, and single cell data analysis software (SeekSoul Tools). Based on the principle of microfluidic technology, the kit uses emulsion droplets to separate and capture individual cells, and then recruits nucleic acid-modified Barcoded Beads to molecularly label RNA from different cell sources. Finally, a high-throughput single-cell transcriptome library compatible with Illumina and MGI sequencing system will be constructed, which can be used for research on tumors, immunity, cell development, viral infections, drug discovery, and target screening, etc.
|
Product |
Product code |
Specification
|
|
SeekOne® DD Single Cell 3’ Transcriptome-seq Kit, 2 tests/8 tests |
K00202-02/K00202-08 |
8 tests
|
Highlights
1. Explore the expression of thousands of genes at the single cell level to gain new insights into cell states and subtypes.
2. Investigate the role of cellular heterogeneity in the biological system at the single cell level.
3. Detect differentially expressed genes between different cell populations to find significant genes, investigate relevant pathways, and explore cellular phenotypes.
4. Analyze cell dynamic changes in various cellular states associated with a continuous process, such as differentiation, by performing trajectory analysis.
5. Address a variety of research needs in cancer, immunity, cell development, viral infection, drug discovery, and target screening.
Applications
Medical and Life Sciences:
1. Construction of large-scale single cell atlases
2. Identification of cell subpopulations/intermediate states/rare cell types
3. Disease heterogeneity/microenvironment/drug resistance/progression
4. Immune cell atlas and differentiation
5. Reproduction/organic neuron/stem cell differentiation and development
6. Disease typing/biomarker discovery
Agronomy:
1. Construction of cell atlas
2. Cell differentiation and development
3. Environmental stress, virus control and other mechanisms
Color blocks are a simple and effective way to present and highlight your content. Choose an image or a color for the background. You can even resize and duplicate the blocks to create your own layout. Add images or icons to customize the blocks.

Workflow
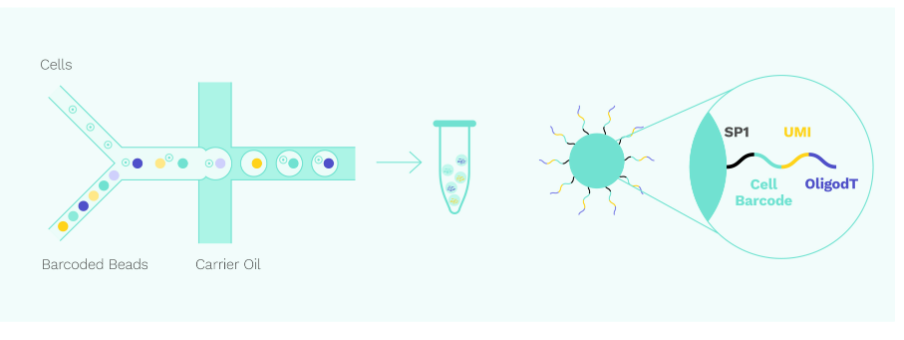
1. Single cell capture and labelling
Cells and barcoded beads are added separately and then react in the carrier oil to form emulsion droplets in the - shaped channel. After that, mRNA molecules released by the cells are captured by oligo(dT) on the barcoded beads.
shaped channel. After that, mRNA molecules released by the cells are captured by oligo(dT) on the barcoded beads.
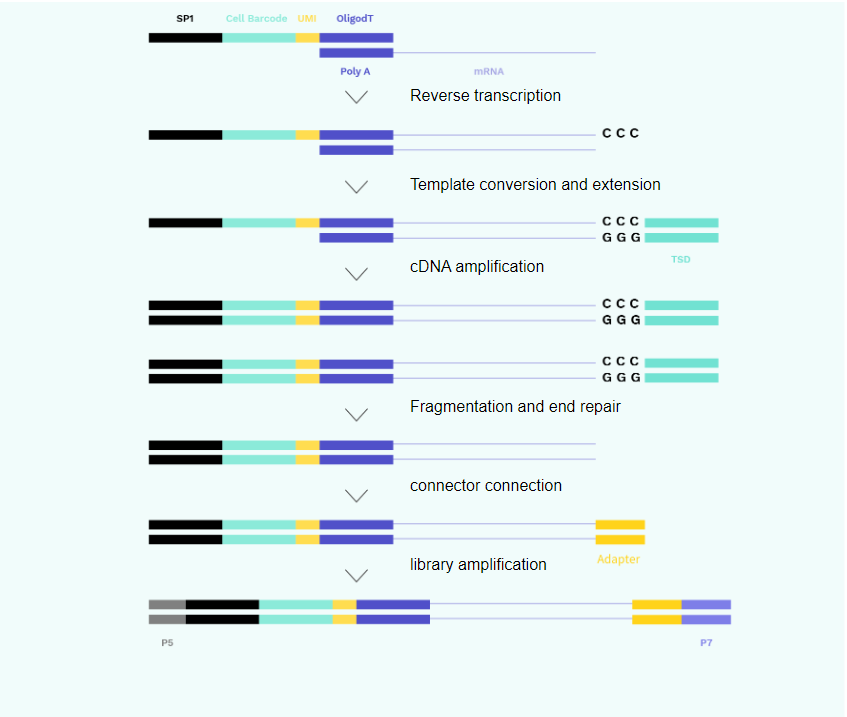
2. Library Construction
Product Features
Compatible
Compatible with single cell 3' transcriptome sequencing (3' scRNA-seq) and single nucleus RNA sequencing (snRNA-seq).
Dual index libraries compatible with both Illumina and MGI sequencers.
Fast
Rapid generation of 150,000 water-in-oil droplets in 3 minutes with high success rates, especially for precious and sensitive samples.
Cost-saving
Unique place chip design eliminates chip waste, reducing operation time and cost
Suitable sample types
a. Delicate samples with low cell viability:brain tissue, retinal tissue, all tissues from aged individuals
b. Tissues requiring nuclei isolation: frozen tissues, pancreatic tissues, neurons, adipose tissue, etc.
c. Large cohort or atlas studies where multiple samples need to be processed simultaneously.
Product Specifications
- Rapidgenerationof150,000water-in-oildropletsin3minutes
- Efficientlycapturing500-12,000cellsperchannel
- Flexiblerunningof1~8samplesinparallel
- Cellsizeflexibility:celldiameterof5~40μm
- Highcellcaptureratesofupto65%
- Low doubletrates ofunder0.3%per1,000cells
Data Presentation
|
Sample Type |
Estimated Number of Cells |
Mean Reads per Cell |
Median Genes per Cell |
Number of Reads |
Valid Barcodes |
Sequencing Saturationn |
Fraction reads in cells |
Total Genes Detected |
Median UMI Counts per Cell |
|
Human-Fresh PBMCs |
1,085 |
19,3061 |
2,116 |
209,664,853 |
94.54% |
92.14% |
77.19% |
18,705 |
7,272 |
|
Human-Glioma |
9,475 |
10,986 |
2,226 |
104,092,350 |
95.07% |
30.26% |
89.99% |
26,004 |
4,527 |
|
Mouse- Subcutaneously Transplanted Tumor |
9,506 |
37,866 |
2,906 |
359,953,834 |
95.99% |
44.78% |
82.99% |
21,558 |
7,563 |
|
Mouse- Frozen Brain Tissue |
9,025 |
9,848 |
998 |
88,873,704 |
94.65% |
90.99% |
76.22% |
22,113 |
1,521 |
|
Human-Adipose Tissue |
12,106 |
24880 |
1,122 |
301,197,280 |
93.89% |
91.28% |
75.54% |
22,334 |
1,954 |