Pyrophosphatase Inorganic (yeast)
Introduction
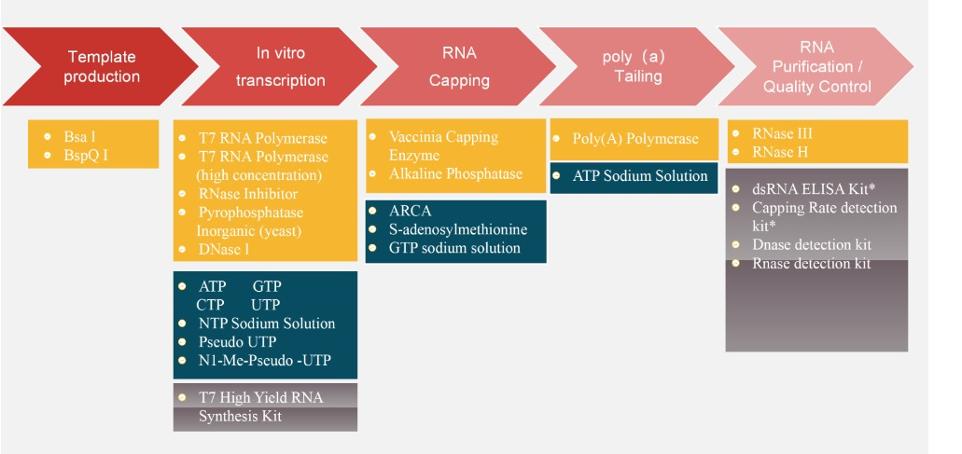
"Gentaur Molecular" operates five major technical platforms concurrently, supporting a cutting-edge special enzyme research and development as well as production system. We offer:
- A complete range of mRNA raw materials and various mRNA stock solutions, continually optimizing and enhancing our products, with key quality indicators of synthetic products at the forefront.
- End-to-end CRO services, encompassing template production, in vitro transcription, capping and tailing, LNP encapsulation, and quality control.
- Comprehensive and top-notch mRNA quality inspection platforms, supported by methods such as capillary electrophoresis, high-performance liquid chromatography (HPLC), liquid chromatography-mass spectrometry (LC-MS), and more."
Our Collaboration
what is Pyrophosphatase Inorganic?
Pyrophosphatase Inorganic (yeast) is derived from a recombinant E. coli strain carrying Saccharomyces cerevisiae ppa gene. The enzyme catalyzes the hydrolysis of inorganic pyrophosphate to form orthophosphate.
P207–4 + H20 → 2HP04–2
Pyrophosphatase Inorganic (yeast) could hydrolyze the inorganic pyrophosphate generated by nucleic acid amplification, in vitro transcription and other reactions, removing the inhibition of the inorganic pyrophosphate generated on the reaction system. The removal of pyrophosphate can shift the reaction equilibrium to the end of product formation, which is beneficial to increase the yield of synthetic product. RNA and DNA synthesis are examples of reactions that can be pulled far in the synthesis direction by the action of inorganic pyrophosphatase.
Pyrophosphatase Inorganic (yeast)
| Catalog | Size |
| HBP000501 | 10U/0.1mL |
| HBP000502 | 50U/0.5mL |
| HBP000503 | 1KU/10mL |
| HBP000504 | 10KU/100mL |

Storage
Store at -20 ℃, valid for 2 years( Avoid repeated freeze-thaw cycles)
Storage Buffer
20 mM Tris-HCl (pH 8.0), 100 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol,50% glycerol。
Unit Definition
One unit is the amount of enzyme that will generate 1 μmol of phosphate per minute from inorganic pyrophosphate under standard reaction conditions. Standard reaction conditions: a 10-minute reaction at 25°C in 100 mM Tris-HCl, [pH 7.2], 2 mM MgCl2 and 2 mM PPi in a reaction volume of 0.5 ml.
Molecular Weight
The molecular weight of Pyrophosphatase, Inorganic (yeast) is 71 kDa.
Quality Control
Exonuclease: 0.1 U of Pyrophosphatase, Inorganic (yeast) with 1 μg λ-Hind III digest DNA at 37 ℃ for 16 hours yields no degradation as determined by agarose gel electrophoresis.
Endonuclease: 0.1 U of Pyrophosphatase, Inorganic (yeast) with 1 μg λDNA at 37 ℃ for 16 hours yields no degradation as determined by agarose gel electrophoresis.
Nickase: 0.1 U of Pyrophosphatase, Inorganic (yeast) with 1 μg pBR322 at 37 ℃ for 16 hours yields no degradation as determined by agarose gel electrophoresis.
RNase: 0.1 U of Pyrophosphatase, Inorganic (yeast) with 1.6 μg MS2 RNA for 4 hours at 37 ℃ yields no degradation as determined by agarose gel electrophoresis.
E. coli DNA: 0.1 U of Pyrophosphatase, Inorganic (yeast) is screened for the presence of E.coli genomic DNA using TaqMan qPCR with primers specific for the E. coli 16S rRNA locus. The E. coli genomic DNA contamination is ≤ 0.1 pg/ 0.1U.
Bacterial Endotoxin: LAL-test, according to Chinese Pharmacopoeia IV 2020 edition, gel limit test method, general rule (1143). Bacterial endotoxin content should be ≤10 EU/mg.
Note on use
- This product is active in a variety of reaction buffers, usually this product can be directly connected in HDA and LAMP amplification experiments.
- It is recommended to optimize the dosage of the enzyme in different experiments, and the concentration is usually 0.05-1 U/mL. 4 U/mL is widely adopted for in vitro transcription system.
- Pyrophosphatase, Inorganic (yeast) is active from 16 to 37°C while the optimum reaction temperature is 25°C.
- For your safety, please wear a lab coat and disposable gloves during operation.
Application
- Enhancing yields of RNA in transcription reactions
Alkaline Phosphatase
Description
Alkaline Phosphatase is derived from a recombinant E. coli strain that carries the TAB5 gene. The enzyme catalyzes the dephosphorylation of 5´ and 3´ ends of DNA and RNA phosphomonoesters. Also, it hydrolyses ribose, as well as deoxyribonucleoside triphosphates (NTPs and dNTPs). TAB5 Alkaline Phosphatase acts on 5´ protruding, 5´ recessed and blunt ends. The Phosphatase can be used in many molecular biology applications, such as cloning or probe end labeling to remove the phosphorylated ends of DNA or RNA. In cloning experiments, dephosphorylation prevents the linearized plasmid DNA from self-ligation. It can also degrade unincorporated dNTPs in PCR reactions to prepare a template for DNA sequencing. The enzyme is completely and irreversibly inactivated by heating at 70°C for 5 minutes, thereby making removal of the phosphatase prior to ligation or end labeling unnecessary.
Alkaline Phosphatase
| Catalog | Size |
| HBP001201 | 0.5KU/0.1mL |
| HBP001202 | 2.5KU/0.5mL |
| HBP001203 | 50KU/10mL |
| HBP001204 | 500KU/100mL |
| HBP001205 | 5000KU/1000mL |

Storage
Store at -20 ℃, valid for 2 year(s Avoid repeated freeze-thaw cycles)
Storage Buffer
10 mM Tris-HCl (pH 7.4,25℃), 1 mM MgCl2 , 0.01 mM ZnCl2, 50% glycerol.
Unit Definition
One unit is defined as the amount of enzyme that will dephosphorylate 1 µg of pUC19 vector DNA cut with HindIII (5´ protruding ends), HincII (blunts ends) or PstI (5´ recessed ends) in 30 minutes at 37°C. Dephosphorylation is defined as > 95% inhibition of recirculation in a self-ligation reaction and is measured by transformation into E.coli.
Molecular Weight
The molecular weight of Alkaline Phosphatase is 35 kDa.
Heat Inactivation
70℃ for 5 minutes
Reaction system and conditions
- Add the following components in the order specified ( Protocol for dephosphorylation of 5´-ends of DNA)
| Components | Dosage |
|---|---|
| DNA | 1 pmol of DNA ends* |
| 10× Reaction Buffer** | 2 µL |
| TAB5 Alkaline Phosphatase | 5 U |
| ddH2O | To 20 μL |
* 1 pmol of DNA ends is about 1 µg of a 3 kb plasmid.
**10× Reaction Buffe : 500 mM Bis-Tris-Propane HCl( pH 6.0,25°C),10 mM MgCl2,1 mM ZnCl2
- Incubate at 37°C for 30 minutes.
- Stop reaction by heat-inactivation at 70°C for 5 minutes.

Quality Control
Exonuclease: 5 U of Alkaline Phosphatase with 1 μg λ-Hind III digest DNA at 37 ℃ for 16 hours yields no degradation as determined by agarose gel electrophoresis.
Endonuclease: 5 U of Alkaline Phosphatase with 1 μg λDNA at 37 ℃ for 16 hours yields no degradation as determined by agarose gel electrophoresis.
Nickase: 5 U of Alkaline Phosphatase with 1 μg pBR322 at 37 ℃ for 16 hours yields no degradation as determined by agarose gel electrophoresis.
RNase: 5 U of Alkaline Phosphatase with 1.6 μg MS2 RNA for 16 hours at 37 ℃ yields no degradation as determined by agarose gel electrophoresis.
E.coli DNA: 5 U of Alkaline Phosphatase is screened for the presence of E. coli genomic DNA using TaqMan qPCR with primers specific for the E. coli 16S rRNA locus. The E. coli genomic DNA contamination is ≤0.1 pg/5 U.
Bacterial Endotoxin: LAL-test, according to Chinese Pharmacopoeia IV 2020 edition, gel limit test method, general rule (1143). Bacterial endotoxin content should be ≤10 EU/mg.

Note on use
- The reaction system can be scaled up or down according to the experimental demands.
- The enzyme is homologous dimer, and its monomer molecular weight is 35 kDa.
- It is indispensable to involve Zn2+ in the reaction process since it is a Zn2+ or Mg2+ dependent enzyme.
- For your safety, please wear lab coat and disposable gloves during operation.
Application
- Dephosphorylation of DNA and RNA
- Prevention of recircularization of cloning vectors
- Removal of dNTPs and pyrophosphate from PCR reactions
- Preparation of templates for 5´ end labeling


